Background Information
What is a G-quadruplex?
The quadruplex structures formed by guanine rich nucleic acid sequences have received significant attention recently because of increasing evidence for their role in important biological processes and as therapeutic targets. The G-quadruplex structure is formed by repeated folding of either the single polynucleotide molecule or by association of two or four molecules. The structure consists of stacked G-tetrads, which are square co-planar arrays of four guanine bases each. G-quadruplexes are stabilized with cyclic Hoogsteen hydrogen bonding between the four guanines within each tetrad.

Significance of G-quadruplexes
G-quadruplex sequence motifs have been reported in telomeric, promoter and other regions of mammalian genomes. G-quadruplex DNA has been suggested to regulate DNA replication and may control cellular proliferation. Although initially most of the studies focused on G-quadruplexes in the DNA, lately there have been many efforts to study G-quadruplex forming RNA. In fact, G-rich sequences capable of forming G-quadruplexes in the RNA have been implicated in a variety of important biological activities, such as mRNA turnover, Fragile X Mental Retardation Protein (FMRP) binding, translation initiation as well as repression.
Untranslated regions and G-quadruplexes
Untranslated regions of eukaryotic mRNAs contain motifs that are vital for regulation of gene expression at post-transcriptional levels. Specific interactions between RNA binding proteins and cis- acting elements in 5’- and 3’- UTRs are responsible for regulating essential biological activities, such as mRNA localization, mRNA turnover, and translation efficiency. Much attention has been paid to study the composition of regulatory RNA motifs and the mechanisms of their interactions with the cellular machinery.
G-quadruplex structure has been implicated in a variety of regulatory processes in the cytoplasm, including mRNA turnover via exoribonuclease action, interaction with FMRP, cap independent translation initiation, and translation repression.
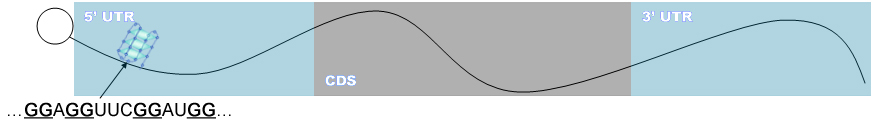
QGRS in the mRNA Eukaryotic mRNA consists of a protein coding sequence, CDS, and 5’ and 3’ untranslated
regions. Regulatory motifs found in these regions are responsible for regulating essential biological activities of the mRNA.
We have performed computational analysis of G-quadruplexes in the untranslated regions of a large number of eukaryotic mRNAs.
The UTR module, an extension of the existing
Quadruplex forming
G-
Rich
Sequences (QGRS) Mapper program (
http://bioinformatics.ramapo.edu/QGRS/), was used for these studies to identify cis- regulatory G-quadruplex motifs in the 5’- and 3’-UTRs. GRS_UTRdb contains data parsed and mapped by the QGRS Mapper.
The database will allow further computation of the data and will help investigate the role of G-quadruplex structures in regulating eukaryotic gene expression.
Related References
- Bagga, P.S., Ford, L.P., Chen, F. and Wilusz, J. (1995) The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3’ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res, 23, 1625-1631.
- Bagga, P.S., Arhin, G.L. and Wilusz, J. (1998) DESF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res, 26, 5343-5350.
- Arhin, G.K., Boots M., Bagga, P.S., Milcarek, C. and Wilusz, J. (2002) Downstream sequence elements with different affinities for the hnRNP H/H’ protein influence the procession efficiency of mammalian polyadenylation signals. Nucleic Acids Res, 30, 1842-1850.
- D’Antonio, L. and Bagga, P.S. (2004) Computational methods for predicting intramolecular G-quadruplexes in nucleotide sequences. Computation Systems Bioinformatics, IEEE: CSB 2004. , 561-562.
- Kostadinov, R., Malhotra, N., Viotti, M., Shine, R., D’Antonio, L. and Bagga, P. (2006) GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res, 34, D119-124.
- Kikin, O., D’Antonio, L., and Bagga, P. (2006) QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res, 34, W676-682.
- Bagga, P. (2007) Bioinformatics approaches for predicting structure and function of UTR. In: Wilusz, J. ed. Methods in Molecular Biology Series: Post-Transcriptional Gene Regulation. The Humana Press Inc., Totowa New Jersey. In Press.
- Kikin, O., Zappala, Z., D'Antonio, L., and Bagga, P. (2007) GRSDB2 and GRS_UTRdb: Databases of Quadruplex Forming G-rich Sequences in pre-mRNAs and mRNAs. Nucleic Acids Research. doi: 10.1093/nar/gkm982.



