GRS UTRdb Help: Search & Analysis
QGRS Definition |
Understanding G-Scores |
Dealing with overlaps |
Search & Analysis |
Glossary
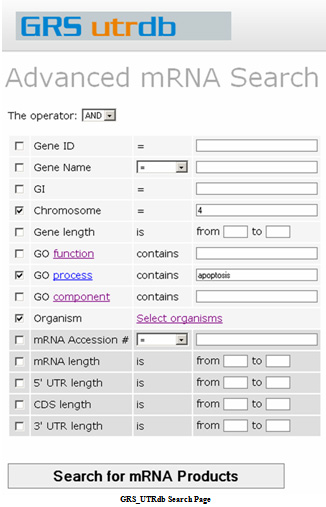
Advanced mRNA Search
The search function contains the following user choices which can be combined with a Boolean operator.(Note: all searches are case-independent.)
Search Field
There are fourteen different fields that the user may search on.
Search Field |
Comments |
Gene ID |
The unique identifier used in Entrez Gene |
Gene Name |
The official NCBI gene name. The user may search for a gene having precisely this name, containing the name, starting or ending with the name. |
GI |
The unique GeneInfo identifier used in NCBI |
Chromosome |
The chromosome on which the gene is located |
Gene length |
A range of values for the length (nucelotides) of the gene |
GO function |
Gene Ontology function |
GO process |
Gene Ontology process |
GO component |
Gene Ontology component |
Organism |
The user may perform a search on a particular set of organisms or all organisms (which is the default). |
mRNA Accession # |
NCBI accession number |
mRNA length |
A range of values for the length of the mRNA product |
5 UTR length |
A range of values for the length of the 5 UTR |
CDS length |
A range of values for the length of the CDS |
3 UTR length |
A range of values for the length of the 3 UTR |
Query results
Suppose we want to analyze human and mouse mRNAs of genes that play a role in RNA binding. Also suppose that we want to examine mRNAs with fairly long 5' and 3' UTRs. We could use the following search parameters
GO function: RNA binding
5' UTR length: 200 to 4000
3' UTR length: 200 to 4000
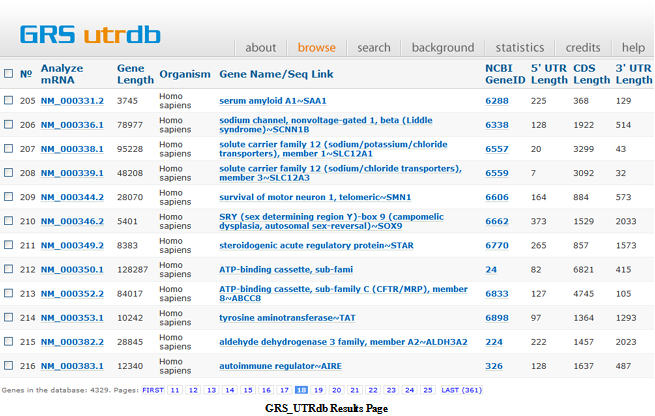
This results in several hits. The first page of results is shown below. The results may be sorted using any of the columns. For example, sorting by Gene Size arranges the genes in increasing order of gene size.
mRNA Analysis
G-quadruplex information may be displayed in six different views:
mRNA Map, Data View (no overlaps), Data View (with overlaps), Sequence View, Gene View, Alternate Products.
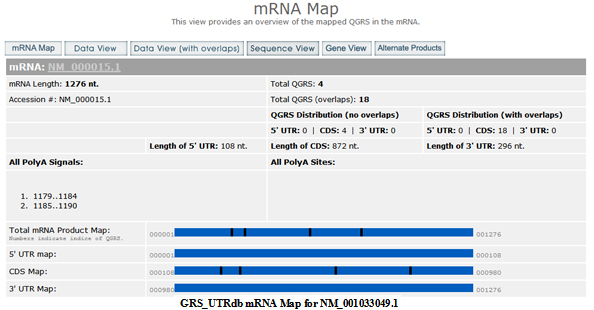
mRNA Map
To analyze a particular mRNA product one clicks on an entry in the
Analyze mRNA column. Suppose we select NM_001033049.1, which corresponds to one of the alternatively spliced products of the ADARB1 (
Adenosine deaminase) gene. This then leads to a page entitled mRNA Map.
This page maps the putative G-quadruplexes (QGRS) in the mRNA product. A table shows an overview of information about the product, including the lengths of the 5 UTR, CDS and 3 UTR. The number of QGRS found in the mRNA is shown. This number is given with and without overlaps. For more information on how overlapping QGRS are handled see the help section
Dealing with Overlaps.
The total number of QGRS is also shown separately for each mRNA section. All known polyA sites and signals are displayed. The page also contains a visual map of the locations of all QGRS in the product.
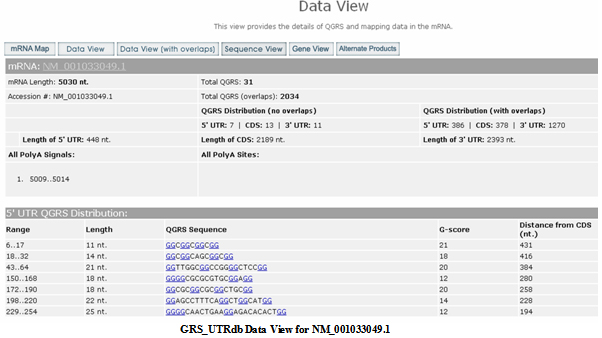
Data View (no overlaps)
Gives the composition and location of each QGRS in 5 UTR, CDS, and 3 UTR for the mRNA product. Along with the specific nucleotide sequence, with G-groups shown in blue and underlined, the view displays the location, length and G-score of the sequence. Each QGRS is assigned a numerical value, called its G-score. The higher the G-score the more likely will the sequence form a stable unimolecular G-quadruplex. For more on G-scores see the help section
Understanding G-Scores.
For the 5 UTR, the distance (nucleotides) from the CDS is given for each QGRS. For the CDS the distance is given to both the initiation and stop codons. For the 3 UTR the distances are given to the CDS, upstream and downstream of the nearest polyA signals, and to the polyA site.
The QGRS displayed in this view are non-overlapping. Please see
Dealing with overlaps to learn about selection of non-overlapping QGRS. Shown below is the Data View for NM_001033049.1, one of the alternatively spliced products of the ADARB1.
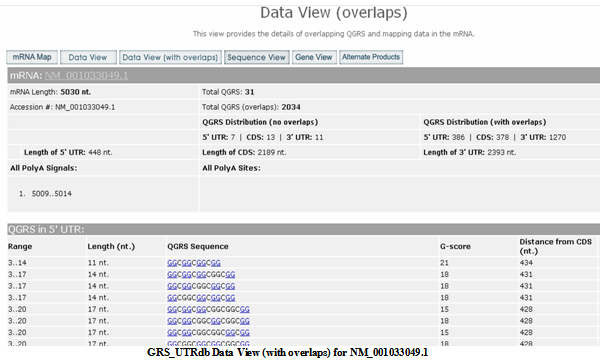
Data View (with overlaps)
Gives a similar display to the above Data View, except that all QGRS, whether overlapping or not, are given.
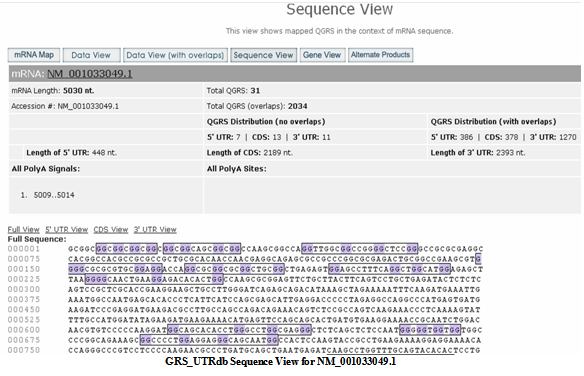
Sequence View
The nucleotide sequence for the entire NM_001033049.1 mRNA product is shown with each QGRS outlined (G-groups shown in purple). The sequences of the 5 UTR, CDS, and 3 UTR are displayed with the corresponding outlined QGRS.
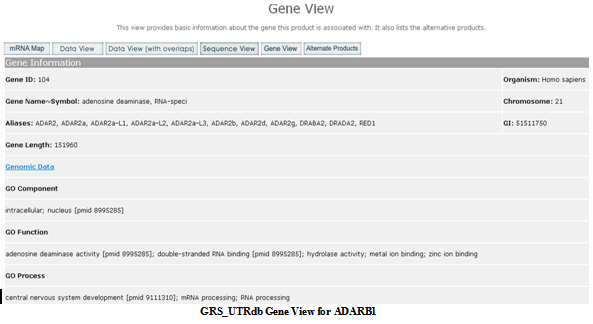
Gene View
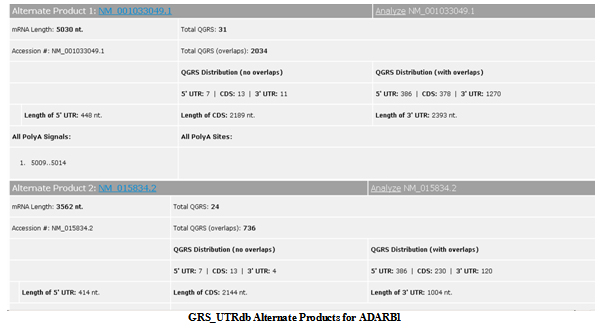
This lists the basic gene information for the mRNA product. A link is given to the NCBI Entrez Gene entry. Also included are all the Gene Ontology terms associated with this gene. Some information is also given for each alternate product associated with the gene and present in the GRS_UTRdb. The information about alternate products can also be accessed directly by the
Alternate Products button at the top of each page.