Background
The ability of G-rich single stranded nucleic acid molecules to form three-dimensional quadruplex structures is well documented
(1,2,3) . The G-quadruplex structure, also known as G-quartet, is composed of stacked G-tetrads, which are square co-planar arrays of four guanine bases each. These interesting structures may be formed by repeated folding of a single nucleic acid molecule or by interaction of two or four strands and are generally very stable due to cyclic Hoogsteen hydrogen bonding between the four guanines within each tetrad.
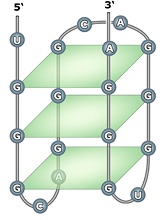
SLC4a3 GRS:
5’ UGGCAGGGCAGGGUGGGA 3’
Predicted intramolecular G-quadruplex
formed by a ‘G’-Rich Sequence (GRS)
found near alternatively spliced site of
cardiac isoform SLC4a3 mouse
transcript.
Naturally occurring G-quadruplex sequence motifs have been reported in telomeric, promoters and other regions of mammalian genomes. ‘G’ rich sequences (GRS) capable of forming G-quadruplexes, have also been implicated in a variety of biological activities such as: mRNA stability (3), transcription pausing (4) , FMRP binding (5) , translation initiation (6) as well as repression (7).
We have previously shown that a conserved ‘G’ rich sequence found in the polyadenylation regions of human genes can mediate efficient 3’end processing of mammalian pre-mRNAs (8,9) , by interacting with DSEF1/hnRNP H’ protein (10) .
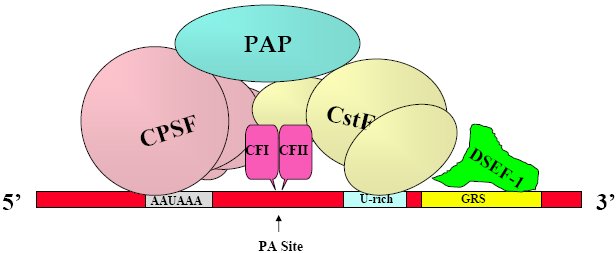
Formation of Cleavage-Polyadenylation complex on mammalian pre-mRNA undergoing 3' end RNA processing
Our preliminary analysis has also revealed the presence of G-rich quadruplex forming sequences near splice junctions of several human transcripts (11) . Members of the hnRNP H protein subfamily, that bind ‘G’ rich motifs, are known to be involved in alternative and tissue specific regulated splicing events (12,13,14) . We believe that G-quadruplexes play a role in modulating the differential RNA processing events by interacting with hnRNP H subfamily of RNA binding proteins.
In order to investigate the role of Quadruplex forming G-Rich Sequences (QGRS) in regulated RNA processing, we have created a suite of computational tools to map putative G-quadruplex elements within mammalian genes. The suite contains algorithms (11) to search genes for occurrences of the G-quadruplex motif and analyze their distribution patterns near RNA processing sites.
QGRS-Mapper:
- Web-based software mainly written in Perl and Java programming languages.
- Retrieves sequence information from fully annotated entries of public genomic databases (e.g. GenBank/RefSeq of NCBI).
- Searches gene sequences for occurrences of Quadruplex forming G-Rich Sequences (QGRS).
- Analyzes their distribution patterns near RNA processing sites.
- Capable of analyzing nucleotide sequences in the raw or FASTA format provided by the user.
GRSDB:
- Web-accessible database for curation and further computation of the mined data.
- Built with PHP/MySQL.
- Stores and organizes the results of the analyses of QGRS-Mapper.
- Helps perform wide-scale analysis of overall occurrence and significance of G-quadruplexes, especially near RNA processing sites.
1 J.T. Davis. Angew. Chem. Int. Ed., 43:668-698. 2004.
2 H. Liu, A. Matsugami, M. Katahira, and S. Uesugi. J. Mol. Biol., 322:955-970, 2002.
3 T. Simonsson. Biol. Chem., 382:621-628, 2001.
4 M. Yonaha and N.J. Proudfoot. Mol. Cell, 3: 593-600, 1999.
5 J.C. Darnell, K.B. Jensen, P. Jin, V. Brown, S.T. Warren, and R.B. Darnell. Cell, 107: 489-499, 2001.
6 S. Bonnal, C. Schaeffer, L. Creancier, S. Clamens, H. Moine, A-C. Prats, and S. Vagner. J Biol. Chem., 278:39330-39336, 2003.
7 A. Oliver, I. Bogdarina, E. Schroeder, I.A. Taylor, and G.G. Kneale. J. Molec. Biol., 301:575-584, 2000.
8 P.S. Bagga, L.C. Ford, F. Chen and J. Wilusz. Nucleic Acids Research. 23:1625-1631, 1995.
9 P.S. Bagga, G.K. Arhin, and J. Wilusz, J. Nucleic Acids Res. 26: 5343-5350, 1998.
10 G.K. Arhin, M. Boots, P.S. Bagga, C. Milcarek, and J. Wilusz. Nucleic Acids Res. 30: 1842-1850, 2002.
11 L. D’Antonio and P.S. Bagga. Computational Systems Bioinformatics, CSB 2004. Proceedings. 2004 IEEE , Pages:561-562. 2004.
12 Min, H., Chan, R.C. and Black, D.L. (1995).Genes Dev., 9: 2659-2671.
12 M.-Y. Chou, N. Rooke, C.W. Turck, and D.L Black. Mol. Cell. Biol. 19: 69-77. 1999.
14 M. Caputi, and A.M. Zahler. EMBO J. 21: 845-855, 2002.






