
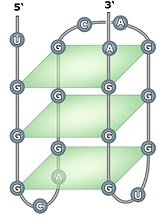
The quadruplex structures formed by guanine rich nucleic acid sequences have received significant attention recently because of increasing evidence for their role in important biological processes and as therapeutic targets. The G-quadruplex structure formed by the repeated folding of either the single polynucleotide molecule or by association of two or four molecules. The structure consists of stacked G-tetrads, which are square co-planar arrays of four guanine bases each. G-quadruplex is stabilized with cyclic Hoogsteen hydrogen bonding between the four guanines within each tetrad.
G-quadruplex sequence motifs have been reported in telomeric, promoter and other regions of mammalian genomes. G-quadruplex DNA has been suggested to regulate DNA replication and may
control cellular proliferation. Although initially most of the studies focused on G-quadruplexes in the DNA, lately there have been many efforts to study G-quadruplexes forming RNA. In fact, G-rich sequences capable of forming G-quadruplexes in the RNA have been implicated in a variety of important biological activities, such as mRNA turnover, Fragile X Mental Retardation Protein (FMRP) binding, translation initiation as well as repression.
We have previously shown that a conserved auxiliary G-rich sequence (GRS) found near the polyadenylation regions can mediate efficient 3’ end processing of mammalian pre-mRNA by interacting with DSEF1/hnRNP H/H’ protein. Regulated polyadenylation is an important component of differential gene expression. An interplay among GRS-binding proteins helps in regulating alternative polyadenylation of mammalian pre-mRNAs. Members of the hnRNP H protein subfamily, that bind G-rich motifs, are also known to be involved in alternative, tissue-specific, regulated splicing events. GRS motifs that are present near splice sites act as splicing regulators by interacting with hnRNP H. The regulatory G-rich motifs may be capable of forming quadruplex structures. Whether quadruplex structure directly plays a role in regulating RNA processing events requires investigation.
Although prevalence of G-quadruplexes in the human genome has been established, there is a paucity of systematic studies focusing on the analysis of G-quadruplex motifs near RNA processing sites, especially those that are alternatively processed. Our group has been interested in studying the role of G-quadruplexes in regulation of gene expression post-transcriptional level. We have adopted a bioinformatics approach to study composition and patterns of G-quadruplexes in pre-mRNA sequences. Our computational suite consists of a "QGRS Mapper" that can analyze genomic nucleotide sequences, and the "GRSDB" and GRS_UTRdb databases for curation and analysis of the QGRS Mapper generated data.
We have been using these servers to perform a large analysis of alternatively processed mammalian transcripts. At present, our database contains over three million G-quadruplex motifs mapped to >29,000 eukaryotic genes of which ~8,000 are alternatively processed.